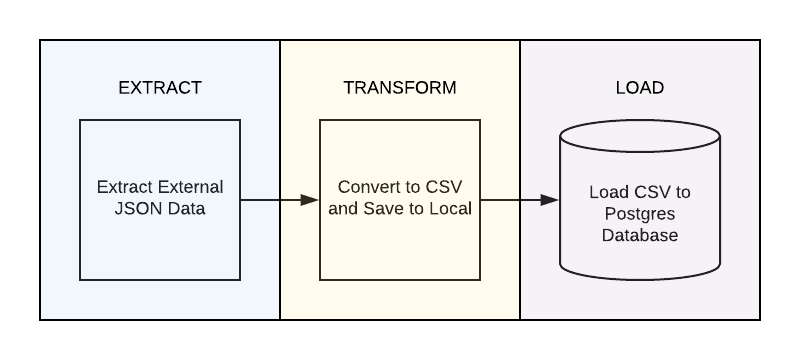
Scenario
We will be using a public open dataset on the counts of COVID-19 related hospitalization, cases, and deaths in New York City as our external data source. The dataset will be pulled as a JSON file and converted to CSV format before being loaded into a PostgreSQL database. Finally, we’ll be using Airflow to orchestrate and automate this pipeline by scheduling the aforementioned steps on a daily basis.
Airflow Installation/ Postgres Setup
Setting up Airflow and an Airflow database is fairly simple but can involve a few steps. For the sake of keeping this article short and focused on Airflow’s scheduling capabilities, please check out this link to setup Postgres and Airflow.
Project Structure
airflow
|---- dags
| |---- covid_dag.py
|---- data
| |---- nyccovid_YYYYMMDD.csv
| |---- nyccovid_YYYYMMDD.csv
|---- logs
|---- airflow.cfg
|---- pg_creds.cfg
|---- unittests.cfg
|---- requirements.txt
Extract
Every pipeline should start with a careful look at the data that will be ingested.
Taking a peek at an example response from the NYC OpenData API, you can see that it shouldn’t be too difficult coming up with a schema for our database.
{
"date_of_interest": "2020-02-29T00:00:00.000",
"case_count": "1",
"hospitalized_count": "14",
"death_count": "0"
},
{
"date_of_interest": "2020-03-01T00:00:00.000",
"case_count": "1",
"hospitalized_count": "4",
"death_count": "0"
},
There are only four columns, all essential for generating meaningful insights. When we get to the transforming stage of our ETL, we could modify the date_of_interest field by dropping the timestamp since it doesn’t look like it is providing any useful information.
Let’s begin our pipeline by creating the covid_data.py file in our airflow/dags directory.
The lone step in the extract stage is to simply request the latest dataset.
url = "https://data.cityofnewyork.us/resource/rc75-m7u3.json"
response = requests.get(url)
Transform
The only modifications that need to be made are converting the JSON to CSV and dropping the timestamp in the date_of_interest field.
Lucky for us, Postgres will implicitly drop the timestamp as long as we cast the date_of_interest column as DATE (instead of TIMESTAMP) when creating the table in our airflow database.
As for the JSON to CSV conversion, we can use the pandas library to load in the JSON response into a pandas DataFrame.
df = pd.DataFrame(json.loads(response.content))
and from there we can easily save the DataFrame as CSV to the airflow/data directory. We will also set the date_of_interest field as the index to avoid any strange formatting issues when saving the DataFrame as a CSV.
df = df.set_index("date_of_interest")
df.to_csv("data/nyccovid_{}.csv".format(date.today().strftime("%Y%m%d")))
Due to the simplicity of this project, we will end up combining the extract and transform sections into one tasks. Obviously for more involved projects, it’s best to break up the pipeline into multiple tasks, or even creating your own custom operators and importing them into your main dag file (don’t worry if that doesn’t make too much sense).
Load
Designing the schema for the airflow database is a must before loading anything into Postgres. The good news is that most the design work was completed during the analysis of the raw data.
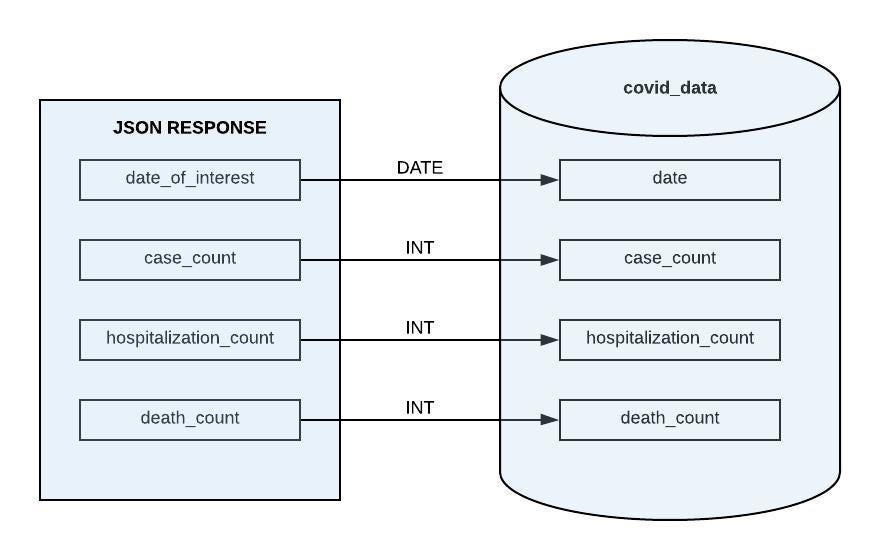
In addition to the actual contents of the data, we need to know what is expected with every new delivery of data.
This is a dataset that is updated daily, and will include old data along with the new. Every dataset contains its entire history so we must be careful to avoid duplicating records when updating our table. A simple solution is to truncate the table before loading new datasets.
Lastly, we should try to avoid hardcoding the database credentials within the covid_data.py script. A better method is to create a separate pg_cred.cfg file in a different directory within the project (I placed mine in airflow/pg_cred.cfg) and use something like ConfigParser to pull that information into our script.
# this will allow us to fetch our credentials from pg_creds.cfg file
config = ConfigParser()
config.read("pg_creds.cfg")
# attempt the connection to postgres
try:
dbconnect = pg.connect(
database=config.get("postgres", "DATABASE"),
user=config.get("postgres", "USERNAME"),
password=config.get("postgres", "PASSWORD"),
host=config.get("postgres", "HOST")
)
except Exception as error:
print(error)
Create a cursor and execute the CREATE TABLE statement containing the appropriate schema
# create the covid_data table that will hold our data and make
# sure to truncate before every load to avoid duplicating rows
cursor = dbconnect.cursor()
cursor.execute("""
CREATE TABLE IF NOT EXISTS covid_data (
date DATE,
case_count INT,
hospitalized_count INT,
death_count INT,
PRIMARY KEY (date)
);
TRUNCATE TABLE covid_data;
"""
)
dbconnect.commit()
Finally, insert each row from our CSV into the covid_data table.
# open the dataset csv file, skip the first row (header), and insert
# each line as a record into the covid_data table
with open("data/nyccovid_{}.csv".format(date.today().strftime("%Y%m%d"))) as f:
next(f)
for row in f:
cursor.execute("""
INSERT INTO covid_data
VALUES ('{}', '{}', '{}', '{}')
""".format(row.split(",")[0], row.split(",")[1], row.split(",")[2], row.split(",")[3])
)
dbconnect.commit()
cursor.close()
dbconnect.close()
Here is a complete look after wrapping our ETL tasks in functions and importing the necessary libraries
Setting Up Our Airflow DAG
Airflow DAGs are composed of tasks created once an operator class is instantiated. In our case, we will be using two PythonOperator classes, one for each ETL function that we previously defined.
To get started, we set the owner and start date (there are many more arguments that can be set) in our default arguments, establish our scheduling interval, and finally define the dependency between tasks using the bit shift operator.
Note the value of “0 1 ” in our schedule_interval argument which is just CRON language for “run daily at 1am”.
from airflow import DAG
from airflow.operators.python_operator import PythonOperator
from datetime import datetime, timedelta
default_args = {
"owner": "airflow",
"start_date": datetime.today() - timedelta(days=1)
}
with DAG(
"covid_nyc_data",
default_args=default_args,
schedule_interval = "0 1 * * *",
) as dag:
fetchDataToLocal = PythonOperator(
task_id="fetch_data_to_local",
python_callable=fetchDataToLocal
)
sqlLoad = PythonOperator(
task_id="sql_load",
python_callable=sqlLoad
)
fetchDataToLocal >> sqlLoad
append this piece of code to the main covid_dag.py script and voila! our ETL/DAG is complete.
Running Our DAG
To test our project, navigate to your terminal and run the following commands
airflow initdb
airflow webserver -p 8080
airflow scheduler
You should be able to access Airflow’s UI by going to your localhost:8080 in your browser. Make sure you toggle the covid_nyc_data DAG on, and click the play button under the links column to immediately trigger the DAG.
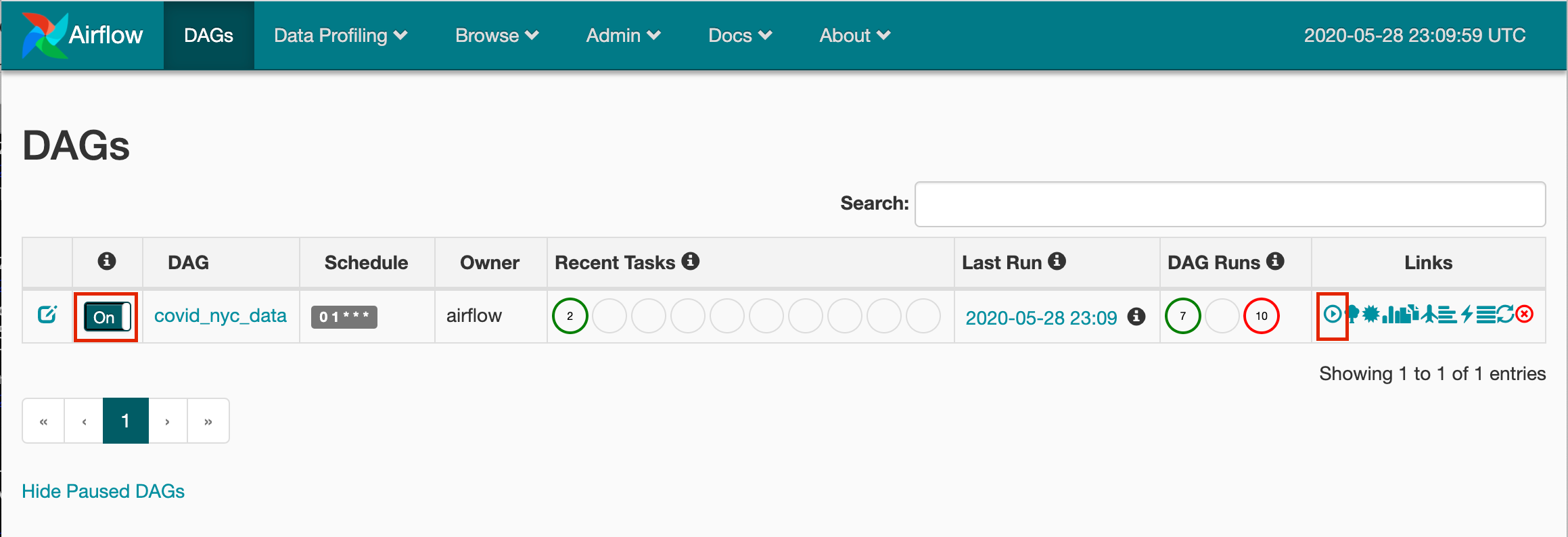
Head over to the Postgres database and perform a SELECT on the covid_data table to verify that our DAG has successfully executed.
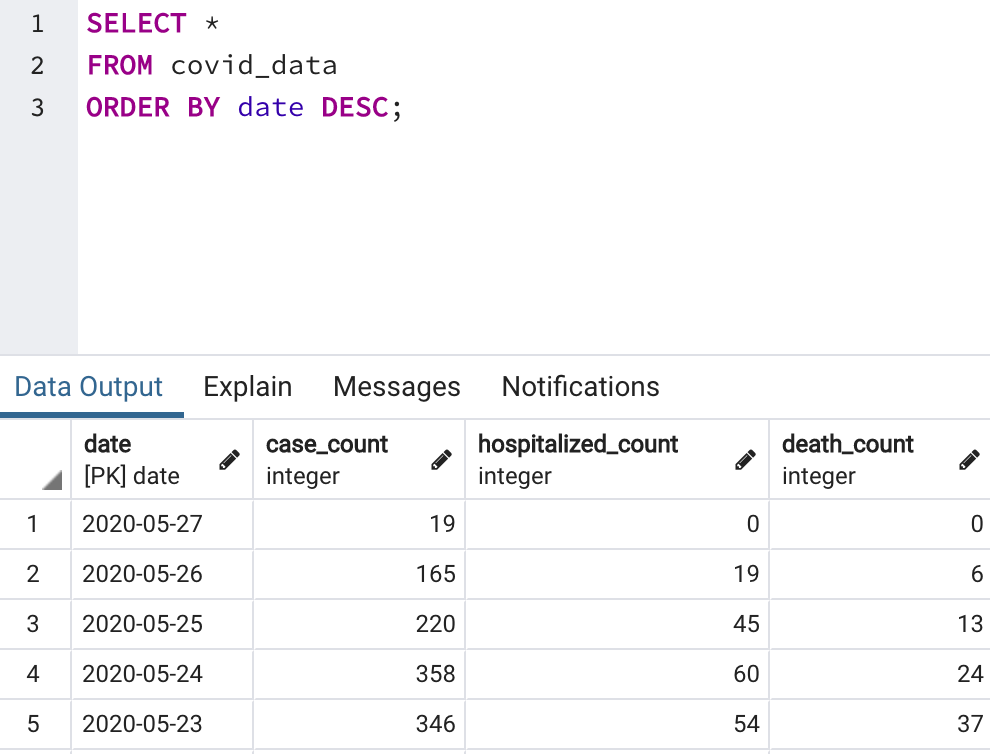
Our pipeline is complete and scheduled to automatically update on a daily basis!
Check out the full repository on my GitHub.
Comments
Loading comments…